function to implement KPCA feature extractor into every model used (LR as first tested model):
widget. You will see explained variance ratio, decision regions, and prediction accuracy of LR model with KPCA extractor as shown in Figure below.
Change data ratio to 0.5 and learning rate 0.1, you will see the decision regions as shown in Figure below.
widget. You will see explained variance ratio, decision regions, and prediction accuracy of perceptron model with KPCA extractor as shown in Figure below.
Change data ratio to 0.4 and learning rate 0.01, you will see the decision regions as shown in Figure below.
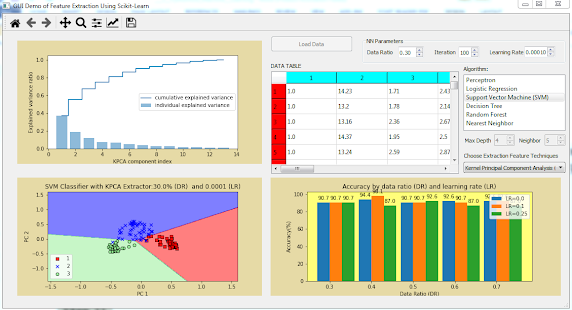
Step 7: Then, you will implement Support Vector Machine (SVM) classifier with KPCA feature extractor. Add the following code to the end of
widget. You will see explained variance ratio, decision regions, and prediction accuracy of SVM model with KPCA extractor as shown in Figure below.
Change data ratio to 0.5 and learning rate 0.5, you will see the decision regions as shown in Figure below.
Step 9: Then, you will implement Decision Tree (DT) classifier with KPCA feature extractor. Add the following code to the end of
widget. You will see explained variance ratio, decision regions, and prediction accuracy of DT model with KPCA extractor as shown in Figure below.
Change data ratio to 0.5 and max depth to 5, you will see the decision regions as shown in Figure below.
Step 11: Then, you will implement Random Forest (RF) classifier with KPCA feature extractor. Add the following code to the end of
widget. You will see explained variance ratio, decision regions, and prediction accuracy of RF model with KPCA extractor as shown in Figure below.
Change data ratio to 0.5 and max depth to 5, you will see the decision regions as shown in Figure below.
Step 13: Lastly, you will implement Nearest Neighbor (KNN) classifier with KPCA feature extractor. Add the following code to the end of
widget. You will see explained variance ratio, decision regions, and prediction accuracy of KNN model with KPCA extractor as shown in Figure below.
Change data ratio to 0.5 and number of neighbors to 7, you will see the decision regions as shown in Figure below.
#Scikit_Classifier_Feature.py
from PyQt5.QtWidgets import *
from PyQt5.uic import loadUi
from matplotlib.backends.backend_qt5agg import (NavigationToolbar2QT as NavigationToolbar)
from matplotlib.colors import ListedColormap
from sklearn import datasets
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split
from sklearn.linear_model import Perceptron
from sklearn.metrics import accuracy_score
from sklearn.linear_model import SGDClassifier
from sklearn.pipeline import make_pipeline
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.decomposition import PCA
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis as LDA
from sklearn.decomposition import KernelPCA as KPCA
from sklearn.linear_model import LogisticRegression
import numpy as np
import pandas as pd
class DemoGUIScikitFeature(QMainWindow):
def __init__(self):
QMainWindow.__init__(self)
loadUi("gui_scikit_feature.ui",self)
self.setWindowTitle("GUI Demo of Feature Extraction Using Scikit-Learn")
self.addToolBar(NavigationToolbar(self.widgetVariance.canvas, self))
self.set_state(False)
self.pbLoad.clicked.connect(self.load_data)
self.listAlgorithm.setCurrentRow(1)
self.listAlgorithm.clicked.connect(self.choose_feature)
self.cboFeature.currentIndexChanged.connect(self.choose_feature)
self.sbIter.valueChanged.connect(self.choose_feature)
self.dsbRatio.valueChanged.connect(self.choose_feature)
self.dsbRate.valueChanged.connect(self.choose_feature)
self.sbDepth.valueChanged.connect(self.choose_feature)
self.sbNeighbor.valueChanged.connect(self.choose_feature)
def set_state(self,state):
self.gbNNParam.setEnabled(state)
self.listAlgorithm.setEnabled(state)
self.sbDepth.setEnabled(state)
self.sbNeighbor.setEnabled(state)
self.cboFeature.setEnabled(state)
def load_data(self):
ratio = self.dsbRatio.value()
self.load_data_ratio(ratio)
#Displays data on table
self.display_table(self.df_wine)
#Enables all parameter widgets
self.set_state(True)
#Disables pbLoad
self.pbLoad.setEnabled(False)
def load_data_ratio(self,ratio):
#Loads wine data
self.df_wine = pd.read_csv(\
'https://archive.ics.uci.edu/ml/machine-learning-
databases/wine/wine.data',header=None)
self.X, self.y = self.df_wine.iloc[:, 1:].values, \
self.df_wine.iloc[:, 0].values
self.X_train, self.X_test, self.y_train, self.y_test = \
train_test_split(self.X, self.y, test_size=ratio, \
stratify=self.y, random_state=0)
#Standardizes the features
sc = StandardScaler()
self.X_train_std = sc.fit_transform(self.X_train)
self.X_test_std = sc.transform(self.X_test)
#Draws explained variance
ylabelStr = 'Explained variance ratio'
xlabelStr = 'Principal component index'
self.draw_exp_var(ylabelStr,xlabelStr,self.widgetVariance.canvas)
def display_table(self,df):
# show data on table widget
self.write_df_to_qtable(df,self.tableData)
styleH = "::section {""background-color: cyan; }"
self.tableData.horizontalHeader().setStyleSheet(styleH)
styleV = "::section {""background-color: red; }"
self.tableData.verticalHeader().setStyleSheet(styleV)
# Takes a df and writes it to a qtable provided. df headers become qtable headers
@staticmethod
def write_df_to_qtable(df,table):
table.setRowCount(df.shape[0])
table.setColumnCount(df.shape[1])
# getting data from df is computationally costly so convert it to array first
df_array = df.values
for row in range(df.shape[0]):
for col in range(df.shape[1]):
table.setItem(row, col, \
QTableWidgetItem(str(df_array[row,col])))
#Displays explained variance ratio
def draw_exp_var(self,ylabelStr,xlabelStr,axisWidget):
#Computes eigenpairs of covariance matrix
cov_mat = np.cov(self.X_train_std.T)
eigen_vals, eigen_vecs = np.linalg.eig(cov_mat)
#Computes total and explained variance
tot = sum(eigen_vals)
var_exp = [(i / tot) for i in sorted(eigen_vals, reverse=True)]
cum_var_exp = np.cumsum(var_exp)
axisWidget.axis1.clear()
axisWidget.axis1.bar(range(1,14), var_exp, alpha=0.5, \
align='center',label='individual explained variance')
axisWidget.axis1.step(range(1,14), cum_var_exp, \
where='mid',label='cumulative explained variance')
axisWidget.axis1.set_ylabel(ylabelStr)
axisWidget.axis1.set_xlabel(xlabelStr)
axisWidget.axis1.legend(loc='best')
axisWidget.draw()
def draw_exp_var_LDA(self,extractor,ylabelStr,xlabelStr,axisWidget):
eigen_vals, eigen_vecs = np.linalg.eig(extractor.covariance_)
tot = sum(eigen_vals.real)
discr = [(i / tot) for i in sorted(eigen_vals.real, reverse=True)]
cum_discr = np.cumsum(discr)
axisWidget.axis1.clear()
axisWidget.axis1.bar(range(1,14), discr, alpha=0.5, \
align='center',label='individual explained variance')
axisWidget.axis1.step(range(1,14), cum_discr, \
where='mid',label='cumulative explained variance')
axisWidget.axis1.set_ylabel(ylabelStr)
axisWidget.axis1.set_xlabel(xlabelStr)
axisWidget.axis1.legend(loc='best')
axisWidget.draw()
def choose_feature(self):
strCB = self.cboFeature.currentText()
if strCB == 'Principal Component Analysis (PCA)':
self.pca_feature()
if strCB == 'Linear Discriminant Analysis (LDA)':
self.lda_feature()
if strCB == 'Kernel Principal Component Analysis (KPCA)':
self.kpca_feature()
def kpca_feature(self):
iterNum = self.sbIter.value()
self.dsbRate.setDecimals(5)
learningRate = self.dsbRate.value()
depth = self.sbDepth.value()
neighbor = self.sbDepth.value()
ratio = self.dsbRatio.value()
self.load_data_ratio(ratio)
item = self.listAlgorithm.currentItem()
strList = item.text()
if strList == 'Logistic Regression':
self.sbIter.setEnabled(True)
self.dsbRate.setEnabled(True)
self.sbDepth.setEnabled(False)
self.sbNeighbor.setEnabled(False)
self.dsbRatio.setEnabled(True)
#Trains LR model
kpca = KPCA(n_components=2, kernel='rbf')
lr = make_pipeline(StandardScaler(), \
SGDClassifier('log',max_iter=iterNum,eta0=learningRate, \
tol=1e-3))
self.X_train_kpca = kpca.fit_transform(self.X_train_std,\
self.y_train)
self.X_test_kpca = kpca.transform(self.X_test_std)
lr.fit(self.X_train_kpca, self.y_train)
#Draws explained variance
ylabelStr = 'Explained variance ratio'
xlabelStr = 'KPCA component index'
self.draw_exp_var(ylabelStr,xlabelStr,self.widgetVariance.canvas)
#Displays decisions region
strTitle = 'LR Classifier with KPCA Extractor: ' + \
str(ratio*100) + '% Data Ratio '
strTitle += ' and Learning Rate ' +str(learningRate)
self.display_decision(self.X_train_kpca, self.y_train, \
classifier=lr,axisWidget=self.widgetDecision.canvas,\
title=strTitle)
#display accuracy graph
self.graph_LR(self.widgetEpoch.canvas, self.accuracy_LR)
if strList == 'Perceptron':
self.sbIter.setEnabled(True)
self.dsbRate.setEnabled(True)
self.sbDepth.setEnabled(False)
self.sbNeighbor.setEnabled(False)
self.dsbRatio.setEnabled(True)
#Trains perceptron
kpca = KPCA(n_components=2, kernel='rbf')
ppn = Perceptron(max_iter=iterNum, eta0=learningRate, \
random_state=1)
self.X_train_kpca = kpca.fit_transform(self.X_train_std,\
self.y_train)
self.X_test_kpca = kpca.transform(self.X_test_std)
ppn.fit(self.X_train_kpca, self.y_train)
#Draws explained variance
ylabelStr = 'Explained variance ratio'
xlabelStr = 'KPCA component index'
self.draw_exp_var(ylabelStr,xlabelStr,self.widgetVariance.canvas)
#Displays decision regions
strTitle = 'Perceptron Classifier with KPCA Extractor: ' + \
str(ratio*100) + '% (DR) '
strTitle += ' and ' +str(learningRate) + ' (LR)'
self.display_decision(self.X_train_kpca, self.y_train, \
classifier=ppn,axisWidget=self.widgetDecision.canvas,\
title=strTitle)
#display graph
self.graph_LR(self.widgetEpoch.canvas, self.accuracy_PPN)
if strList == 'Support Vector Machine (SVM)':
self.sbIter.setEnabled(True)
self.dsbRate.setEnabled(True)
self.sbDepth.setEnabled(False)
self.sbNeighbor.setEnabled(False)
self.dsbRatio.setEnabled(True)
#Trains SVM model
kpca = KPCA(n_components=2, kernel='rbf')
svm = make_pipeline(StandardScaler(), \
SGDClassifier('hinge',max_iter=iterNum,eta0=learningRate, \
tol=1e-3))
self.X_train_kpca = kpca.fit_transform(self.X_train_std,\
self.y_train)
self.X_test_kpca = kpca.transform(self.X_test_std)
svm.fit(self.X_train_kpca, self.y_train)
#Draws explained variance
ylabelStr = 'Explained variance ratio'
xlabelStr = 'KPCA component index'
self.draw_exp_var(ylabelStr,xlabelStr,self.widgetVariance.canvas)
#Displays decision regions
strTitle = 'SVM Classifier with KPCA Extractor:' +\
str(ratio*100) + '% (DR) '
strTitle += ' and ' +str(learningRate) + ' (LR)'
self.display_decision(self.X_train_kpca, self.y_train, \
classifier=svm,axisWidget=self.widgetDecision.canvas,\
title=strTitle)
#display graph
self.graph_LR(self.widgetEpoch.canvas, self.accuracy_SVM)
if strList == 'Decision Tree':
self.dsbRatio.setEnabled(True)
self.sbIter.setEnabled(False)
self.dsbRate.setEnabled(False)
self.sbDepth.setEnabled(True)
self.sbIter.setEnabled(False)
self.sbNeighbor.setEnabled(False)
#Trains Decision Tree model
kpca = KPCA(n_components=2, kernel='rbf')
tree = DecisionTreeClassifier(criterion='gini', \
max_depth=depth,random_state=1)
self.X_train_kpca = kpca.fit_transform(self.X_train_std,\
self.y_train)
self.X_test_kpca = kpca.transform(self.X_test_std)
tree.fit(self.X_train_kpca, self.y_train)
#Draws explained variance
ylabelStr = 'Explained variance ratio'
xlabelStr = 'KPCA component index'
self.draw_exp_var(ylabelStr,xlabelStr,self.widgetVariance.canvas)
#Displays decision regions
strTitle = 'DT Classifier with KPCA Extractor: (DR)=' + \
str(ratio*100)
strTitle += ' and Max Depth=' +str(depth)
self.display_decision(self.X_train_kpca, self.y_train, \
classifier=tree,axisWidget=self.widgetDecision.canvas,\
title=strTitle)
#display accuracy graph
self.graph_DT(self.widgetEpoch.canvas, self.accuracy_DT)
if strList == 'Random Forest':
self.dsbRatio.setEnabled(True)
self.sbIter.setEnabled(False)
self.dsbRate.setEnabled(False)
self.sbDepth.setEnabled(True)
self.sbIter.setEnabled(False)
self.sbNeighbor.setEnabled(False)
#Trains Random Forest model
kpca = KPCA(n_components=2, kernel='rbf')
forest = RandomForestClassifier(criterion='gini', \
n_estimators=25,max_depth=depth,random_state=1)
self.X_train_kpca = kpca.fit_transform(self.X_train_std,\
self.y_train)
self.X_test_kpca = kpca.transform(self.X_test_std)
forest.fit(self.X_train_kpca, self.y_train)
#Draws explained variance
ylabelStr = 'Explained variance ratio'
xlabelStr = 'KPCA component index'
self.draw_exp_var(ylabelStr,xlabelStr,self.widgetVariance.canvas)
#Draws explained variance
strTitle = 'Random Forest Classifier with (DR)=' + str(ratio*100)
strTitle += ' and Max Depth =' +str(depth)
self.display_decision(self.X_train_kpca, self.y_train, \
classifier=forest,axisWidget=self.widgetDecision.canvas,\
title=strTitle)
#display accuracy graph
self.graph_DT(self.widgetEpoch.canvas, self.accuracy_RF)
if strList == 'Nearest Neighbor':
self.dsbRatio.setEnabled(True)
self.sbIter.setEnabled(False)
self.dsbRate.setEnabled(False)
self.sbDepth.setEnabled(False)
self.sbIter.setEnabled(False)
self.sbNeighbor.setEnabled(True)
#Trains Nearest Neighbor model
kpca = KPCA(n_components=2, kernel='rbf')
knn = KNeighborsClassifier(n_neighbors=neighbor, p=2, \
metric='minkowski')
self.X_train_kpca = kpca.fit_transform(self.X_train_std,\
self.y_train)
self.X_test_kpca = kpca.transform(self.X_test_std)
knn.fit(self.X_train_kpca, self.y_train)
#Draws explained variance
ylabelStr = 'Explained variance ratio'
xlabelStr = 'KPCA component index'
self.draw_exp_var(ylabelStr,xlabelStr,self.widgetVariance.canvas)
strTitle = 'KNN Classifier with (DR)=' + str(ratio*100)
strTitle += ' and Neihbors =' +str(neighbor)
self.display_decision(self.X_train_kpca, self.y_train, \
classifier=knn,axisWidget=self.widgetDecision.canvas,\
title=strTitle)
#display accuracy graph
self.graph_KNN(self.widgetEpoch.canvas, self.accuracy_KNN)
def lda_feature(self):
iterNum = self.sbIter.value()
self.dsbRate.setDecimals(5)
learningRate = self.dsbRate.value()
depth = self.sbDepth.value()
neighbor = self.sbDepth.value()
ratio = self.dsbRatio.value()
self.load_data_ratio(ratio)
item = self.listAlgorithm.currentItem()
strList = item.text()
if strList == 'Logistic Regression':
self.sbIter.setEnabled(True)
self.dsbRate.setEnabled(True)
self.sbDepth.setEnabled(False)
self.sbNeighbor.setEnabled(False)
self.dsbRatio.setEnabled(True)
#Trains LR model
lda = LDA(n_components=2, store_covariance=True)
lr = make_pipeline(StandardScaler(), \
SGDClassifier('log',max_iter=iterNum,eta0=learningRate, \
tol=1e-3))
self.X_train_lda = lda.fit_transform(self.X_train_std,\
self.y_train)
self.X_test_lda = lda.transform(self.X_test_std)
lr.fit(self.X_train_lda, self.y_train)
#Draws explained variance
ylabelStr = 'Explained variance ratio'
xlabelStr = 'LDA component index'
self.draw_exp_var_LDA(lda,ylabelStr,xlabelStr,\
self.widgetVariance.canvas)
#Displays decisions region
strTitle = 'LR Classifier with LDA Extractor: ' + \
str(ratio*100) + '% Data Ratio '
strTitle += ' and Learning Rate ' +str(learningRate)
self.display_decision(self.X_train_lda, self.y_train,\
classifier=lr,axisWidget=self.widgetDecision.canvas,\
title=strTitle)
#display accuracy graph
self.graph_LR(self.widgetEpoch.canvas, self.accuracy_LR)
if strList == 'Perceptron':
self.sbIter.setEnabled(True)
self.dsbRate.setEnabled(True)
self.sbDepth.setEnabled(False)
self.sbNeighbor.setEnabled(False)
self.dsbRatio.setEnabled(True)
#Trains perceptron
lda = LDA(n_components=2, store_covariance=True)
ppn = Perceptron(max_iter=iterNum, eta0=learningRate, \
random_state=1)
self.X_train_lda = lda.fit_transform(self.X_train_std,\
self.y_train)
self.X_test_lda = lda.transform(self.X_test_std)
ppn.fit(self.X_train_lda, self.y_train)
#Draws explained variance
ylabelStr = 'Explained variance ratio'
xlabelStr = 'LDA component index'
self.draw_exp_var_LDA(lda,ylabelStr,xlabelStr,\
self.widgetVariance.canvas)
#Displays decision regions
strTitle = 'Perceptron Classifier with LDA Extractor: ' + \
str(ratio*100) + '% (DR) '
strTitle += ' and ' +str(learningRate) + ' (LR)'
self.display_decision(self.X_train_lda, self.y_train, \
classifier=ppn,axisWidget=self.widgetDecision.canvas,\
title=strTitle)
#display graph
self.graph_LR(self.widgetEpoch.canvas, self.accuracy_PPN)
if strList == 'Support Vector Machine (SVM)':
self.sbIter.setEnabled(True)
self.dsbRate.setEnabled(True)
self.sbDepth.setEnabled(False)
self.sbNeighbor.setEnabled(False)
self.dsbRatio.setEnabled(True)
#Trains SVM model
lda = LDA(n_components=2, store_covariance=True)
svm = make_pipeline(StandardScaler(), \
SGDClassifier('hinge',max_iter=iterNum,eta0=learningRate, \
tol=1e-3))
self.X_train_lda = lda.fit_transform(self.X_train_std,\
self.y_train)
self.X_test_lda = lda.transform(self.X_test_std)
svm.fit(self.X_train_lda, self.y_train)
#Draws explained variance
ylabelStr = 'Explained variance ratio'
xlabelStr = 'LDA component index'
self.draw_exp_var_LDA(lda,ylabelStr,xlabelStr,\
self.widgetVariance.canvas)
#Displays decision regions
strTitle = 'SVM Classifier with PCA Extractor:' + \
str(ratio*100) + '% (DR) '
strTitle += ' and ' +str(learningRate) + ' (LR)'
self.display_decision(self.X_train_lda, self.y_train, \
classifier=svm,axisWidget=self.widgetDecision.canvas,\
title=strTitle)
#display graph
self.graph_LR(self.widgetEpoch.canvas, self.accuracy_SVM)
if strList == 'Decision Tree':
self.dsbRatio.setEnabled(True)
self.sbIter.setEnabled(False)
self.dsbRate.setEnabled(False)
self.sbDepth.setEnabled(True)
self.sbIter.setEnabled(False)
self.sbNeighbor.setEnabled(False)
#Trains Decision Tree model
lda = LDA(n_components=2, store_covariance=True)
tree = DecisionTreeClassifier(criterion='gini', \
max_depth=depth,random_state=1)
self.X_train_lda = lda.fit_transform(self.X_train_std,\
self.y_train)
self.X_test_lda = lda.transform(self.X_test_std)
tree.fit(self.X_train_lda, self.y_train)
#Draws explained variance
ylabelStr = 'Explained variance ratio'
xlabelStr = 'LDA component index'
self.draw_exp_var_LDA(lda,ylabelStr,xlabelStr,\
self.widgetVariance.canvas)
#Displays decision regions
strTitle = 'DT Classifier with PCA Extractor: (DR)=' + \
str(ratio*100)
strTitle += ' and Max Depth=' +str(depth)
self.display_decision(self.X_train_lda, self.y_train, \
classifier=tree,axisWidget=self.widgetDecision.canvas,\
title=strTitle)
#display accuracy graph
self.graph_DT(self.widgetEpoch.canvas, self.accuracy_DT)
if strList == 'Random Forest':
self.dsbRatio.setEnabled(True)
self.sbIter.setEnabled(False)
self.dsbRate.setEnabled(False)
self.sbDepth.setEnabled(True)
self.sbIter.setEnabled(False)
self.sbNeighbor.setEnabled(False)
#Trains Random Forest model
lda = LDA(n_components=2, store_covariance=True)
forest = RandomForestClassifier(criterion='gini', \
n_estimators=25,max_depth=depth,random_state=1)
self.X_train_lda = lda.fit_transform(self.X_train_std,self.y_train)
self.X_test_lda = lda.transform(self.X_test_std)
forest.fit(self.X_train_lda, self.y_train)
#Draws explained variance
ylabelStr = 'Explained variance ratio'
xlabelStr = 'LDA component index'
self.draw_exp_var_LDA(lda,ylabelStr,xlabelStr,\
self.widgetVariance.canvas)
#Draws explained variance
strTitle = 'Random Forest Classifier with (DR)=' + str(ratio*100)
strTitle += ' and Max Depth =' +str(depth)
self.display_decision(self.X_train_lda, self.y_train, \
classifier=forest,axisWidget=self.widgetDecision.canvas,\
title=strTitle)
#display accuracy graph
self.graph_DT(self.widgetEpoch.canvas, self.accuracy_RF)
if strList == 'Nearest Neighbor':
self.dsbRatio.setEnabled(True)
self.sbIter.setEnabled(False)
self.dsbRate.setEnabled(False)
self.sbDepth.setEnabled(False)
self.sbIter.setEnabled(False)
self.sbNeighbor.setEnabled(True)
#Trains Nearest Neighbor model
lda = LDA(n_components=2, store_covariance=True)
knn = KNeighborsClassifier(n_neighbors=neighbor, p=2, \
metric='minkowski')
self.X_train_lda = lda.fit_transform(self.X_train_std,self.y_train)
self.X_test_lda = lda.transform(self.X_test_std)
knn.fit(self.X_train_lda, self.y_train)
#Draws explained variance
ylabelStr = 'Explained variance ratio'
xlabelStr = 'LDA component index'
self.draw_exp_var_LDA(lda,ylabelStr,xlabelStr,\
self.widgetVariance.canvas)
strTitle = 'KNN Classifier with (DR)=' + str(ratio*100)
strTitle += ' and Neihbors =' +str(neighbor)
self.display_decision(self.X_train_lda, self.y_train, \
classifier=knn,axisWidget=self.widgetDecision.canvas,\
title=strTitle)
#display accuracy graph
self.graph_KNN(self.widgetEpoch.canvas, self.accuracy_KNN)
def pca_feature(self):
iterNum = self.sbIter.value()
self.dsbRate.setDecimals(5)
learningRate = self.dsbRate.value()
depth = self.sbDepth.value()
neighbor = self.sbDepth.value()
ratio = self.dsbRatio.value()
self.load_data_ratio(ratio)
item = self.listAlgorithm.currentItem()
strList = item.text()
if strList == 'Logistic Regression':
self.sbIter.setEnabled(True)
self.dsbRate.setEnabled(True)
self.sbDepth.setEnabled(False)
self.sbNeighbor.setEnabled(False)
self.dsbRatio.setEnabled(True)
pca = PCA(n_components=2)
lr = make_pipeline(StandardScaler(), \
SGDClassifier('log',max_iter=iterNum,eta0=learningRate, \
tol=1e-3))
self.X_train_pca = pca.fit_transform(self.X_train_std)
self.X_test_pca = pca.transform(self.X_test_std)
lr.fit(self.X_train_pca, self.y_train)
strTitle = 'LR Classifier with PCA Extractor: ' + \
str(ratio*100) + '% Data Ratio '
strTitle += ' and Learning Rate ' +str(learningRate)
self.display_decision(self.X_train_pca, self.y_train, \
classifier=lr,axisWidget=self.widgetDecision.canvas,\
title=strTitle)
#display accuracy graph
self.graph_LR(self.widgetEpoch.canvas, self.accuracy_LR)
if strList == 'Perceptron':
self.sbIter.setEnabled(True)
self.dsbRate.setEnabled(True)
self.sbDepth.setEnabled(False)
self.sbNeighbor.setEnabled(False)
self.dsbRatio.setEnabled(True)
#Trains perceptron
pca = PCA(n_components=2)
ppn = Perceptron(max_iter=iterNum, eta0=learningRate, \
random_state=1)
self.X_train_pca = pca.fit_transform(self.X_train_std)
self.X_test_pca = pca.transform(self.X_test_std)
ppn.fit(self.X_train_pca, self.y_train)
strTitle = 'Perceptron Classifier with PCA Extractor: ' + \
str(ratio*100) + '% (DR) '
strTitle += ' and ' +str(learningRate) + ' (LR)'
self.display_decision(self.X_train_pca, self.y_train, \
classifier=ppn,axisWidget=self.widgetDecision.canvas,\
title=strTitle)
#display graph
self.graph_LR(self.widgetEpoch.canvas, self.accuracy_PPN)
if strList == 'Support Vector Machine (SVM)':
self.sbIter.setEnabled(True)
self.dsbRate.setEnabled(True)
self.sbDepth.setEnabled(False)
self.sbNeighbor.setEnabled(False)
self.dsbRatio.setEnabled(True)
#Trains SVM model
pca = PCA(n_components=2)
svm = make_pipeline(StandardScaler(), \
SGDClassifier('hinge',max_iter=iterNum,eta0=learningRate, \
tol=1e-3))
self.X_train_pca = pca.fit_transform(self.X_train_std)
self.X_test_pca = pca.transform(self.X_test_std)
svm.fit(self.X_train_pca, self.y_train)
strTitle = 'SVM Classifier with PCA Extractor:' + \
str(ratio*100) + '% (DR) '
strTitle += ' and ' +str(learningRate) + ' (LR)'
self.display_decision(self.X_train_pca, self.y_train,\
classifier=svm,axisWidget=self.widgetDecision.canvas,\
title=strTitle)
#display graph
self.graph_LR(self.widgetEpoch.canvas, self.accuracy_SVM)
if strList == 'Decision Tree':
self.dsbRatio.setEnabled(True)
self.sbIter.setEnabled(False)
self.dsbRate.setEnabled(False)
self.sbDepth.setEnabled(True)
self.sbIter.setEnabled(False)
self.sbNeighbor.setEnabled(False)
#Trains Decision Tree model
pca = PCA(n_components=2)
tree = DecisionTreeClassifier(criterion='gini',\
max_depth=depth,random_state=1)
self.X_train_pca = pca.fit_transform(self.X_train_std)
self.X_test_pca = pca.transform(self.X_test_std)
tree.fit(self.X_train_pca, self.y_train)
strTitle = 'DT Classifier with PCA Extractor: (DR)=' + \
str(ratio*100)
strTitle += ' and Max Depth=' +str(depth)
self.display_decision(self.X_train_pca, self.y_train, \
classifier=tree,axisWidget=self.widgetDecision.canvas,\
title=strTitle)
#display accuracy graph
self.graph_DT(self.widgetEpoch.canvas, self.accuracy_DT)
if strList == 'Random Forest':
self.dsbRatio.setEnabled(True)
self.sbIter.setEnabled(False)
self.dsbRate.setEnabled(False)
self.sbDepth.setEnabled(True)
self.sbIter.setEnabled(False)
self.sbNeighbor.setEnabled(False)
#Trains Random Forest model
pca = PCA(n_components=2)
forest = RandomForestClassifier(criterion='gini', \
n_estimators=25,max_depth=depth,random_state=1)
self.X_train_pca = pca.fit_transform(self.X_train_std)
self.X_test_pca = pca.transform(self.X_test_std)
forest.fit(self.X_train_pca, self.y_train)
strTitle = 'Random Forest Classifier with (DR)=' + str(ratio*100)
strTitle += ' and Max Depth =' +str(depth)
self.display_decision(self.X_train_pca, self.y_train, \
classifier=forest,axisWidget=self.widgetDecision.canvas,\
title=strTitle)
#display accuracy graph
self.graph_DT(self.widgetEpoch.canvas, self.accuracy_RF)
if strList == 'Nearest Neighbor':
self.dsbRatio.setEnabled(True)
self.sbIter.setEnabled(False)
self.dsbRate.setEnabled(False)
self.sbDepth.setEnabled(False)
self.sbIter.setEnabled(False)
self.sbNeighbor.setEnabled(True)
#Trains Nearest Neighbor model
pca = PCA(n_components=2)
knn = KNeighborsClassifier(n_neighbors=neighbor, p=2, \
metric='minkowski')
self.X_train_pca = pca.fit_transform(self.X_train_std)
self.X_test_pca = pca.transform(self.X_test_std)
knn.fit(self.X_train_pca, self.y_train)
strTitle = 'KNN Classifier with (DR)=' + str(ratio*100)
strTitle += ' and Neihbors =' +str(neighbor)
self.display_decision(self.X_train_pca, self.y_train, \
classifier=knn,axisWidget=self.widgetDecision.canvas,\
title=strTitle)
#display accuracy graph
self.graph_KNN(self.widgetEpoch.canvas, self.accuracy_KNN)
def display_decision(self,X, y, classifier, axisWidget,title,\
resolution=0.01):
# setup marker generator and color map
markers = ('s', 'x', 'o', '^', 'v')
colors = ('red', 'blue', 'lightgreen', 'gray', 'cyan')
cmap = ListedColormap(colors[:len(np.unique(y))])
# plot the decision surface
x1_min, x1_max = X[:, 0].min() - 1, X[:, 0].max() + 1
x2_min, x2_max = X[:, 1].min() - 1, X[:, 1].max() + 1
xx1, xx2 = np.meshgrid(np.arange(x1_min, x1_max, resolution),
np.arange(x2_min, x2_max, resolution))
Z = classifier.predict(np.array([xx1.ravel(), xx2.ravel()]).T)
Z = Z.reshape(xx1.shape)
axisWidget.axis1.clear()
axisWidget.axis1.contourf(xx1, xx2, Z, alpha=0.5, cmap=cmap)
axisWidget.axis1.set_xlim(xx1.min(), xx1.max())
axisWidget.axis1.set_ylim(xx2.min(), xx2.max())
# plot class samples
for idx, cl in enumerate(np.unique(y)):
axisWidget.axis1.scatter(x=X[y == cl, 0],
y=X[y == cl, 1],
alpha=0.8,
c=colors[idx],
marker=markers[idx],
label=cl,
edgecolor='black')
axisWidget.axis1.set_xlabel('PC 1')
axisWidget.axis1.set_ylabel('PC 2')
axisWidget.axis1.legend(loc='lower left')
axisWidget.axis1.set_title(title)
axisWidget.draw()
def accuracy_PPN(self,dataRatio,lRate):
pca = PCA(n_components=2)
ppn = Perceptron(max_iter=1000, eta0=lRate, random_state=1)
self.X_train_pca = pca.fit_transform(self.X_train_std)
self.X_test_pca = pca.transform(self.X_test_std)
ppn.fit(self.X_train_pca, self.y_train)
#Makes prediction
y_pred = ppn.predict(self.X_test_pca)
#Calculates classification accuracy
acc = round(100*accuracy_score(self.y_test, y_pred),1)
return acc
def accuracy_SVM(self,dataRatio,lRate):
pca = PCA(n_components=2)
svm = make_pipeline(StandardScaler(), \
SGDClassifier('hinge',max_iter=1000,eta0=lRate, tol=1e-3))
self.X_train_pca = pca.fit_transform(self.X_train_std)
self.X_test_pca = pca.transform(self.X_test_std)
svm.fit(self.X_train_pca, self.y_train)
#Makes prediction
y_pred = svm.predict(self.X_test_pca)
#Calculates classification accuracy
acc = round(100*accuracy_score(self.y_test, y_pred),1)
return acc
def accuracy_DT(self,ratio,depth):
#Trains Decision Tree model
pca = PCA(n_components=2)
tree = DecisionTreeClassifier(criterion='gini', \
max_depth=depth,random_state=1)
self.X_train_pca = pca.fit_transform(self.X_train_std)
self.X_test_pca = pca.transform(self.X_test_std)
tree.fit(self.X_train_pca, self.y_train)
#Makes prediction
y_pred = tree.predict(self.X_test_pca)
#Calculates classification accuracy
acc = round(100*accuracy_score(self.y_test, y_pred),1)
return acc
def accuracy_RF(self,ratio,depth):
#Trains Decision Tree model
pca = PCA(n_components=2)
forest = RandomForestClassifier(criterion='gini', \
n_estimators=25,max_depth=depth,random_state=1)
self.X_train_pca = pca.fit_transform(self.X_train_std)
self.X_test_pca = pca.transform(self.X_test_std)
forest.fit(self.X_train_pca, self.y_train)
#Makes prediction
y_pred = forest.predict(self.X_test_pca)
#Calculates classification accuracy
acc = round(100*accuracy_score(self.y_test, y_pred),1)
return acc
def accuracy_KNN(self,ratio,neighbor):
pca = PCA(n_components=2)
knn = KNeighborsClassifier(n_neighbors=neighbor, p=2, \
metric='minkowski')
self.X_train_pca = pca.fit_transform(self.X_train_std)
self.X_test_pca = pca.transform(self.X_test_std)
knn.fit(self.X_train_pca, self.y_train)
#Makes prediction
y_pred = knn.predict(self.X_test_pca)
#Calculates classification accuracy
acc = round(100*accuracy_score(self.y_test, y_pred),1)
return acc
def accuracy_LR(self,ratio,lRate):
pca = PCA(n_components=2)
lr = make_pipeline(StandardScaler(), \
SGDClassifier('log',max_iter=1000,eta0=lRate, tol=1e-3))
self.X_train_pca = pca.fit_transform(self.X_train_std)
self.X_test_pca = pca.transform(self.X_test_std)
lr.fit(self.X_train_pca, self.y_train)
#Makes prediction
y_pred = lr.predict(self.X_test_pca)
#Calculates classification accuracy
acc = round(100*accuracy_score(self.y_test, y_pred),1)
return acc
def graph_KNN(self,axisWidget,func):
ratio = self.dsbRatio.value()
neighbor = self.sbNeighbor.value()
if (ratio+0.4) < 1 :
rangeDR = [ratio,ratio+0.1,ratio+0.2,ratio+0.3,ratio+0.4]
else :
rangeDR = [ratio-0.4,ratio-0.3,ratio-0.2,ratio-0.1,ratio]
labels = [str(round(rangeDR[0],2)), str(round(rangeDR[1],2)), \
str(round(rangeDR[2],2)), str(round(rangeDR[3],2)), \
str(round(rangeDR[4],2))]
Neighbor1 = []
for i in rangeDR:
acc = func(i,neighbor)
Neighbor1.append(acc)
Neighbor2 = []
for i in rangeDR:
acc = func(i,neighbor+2)
Neighbor2.append(acc)
Neighbor3 = []
for i in rangeDR:
acc = func(i,neighbor+3)
Neighbor3.append(acc)
x = np.arange(len(labels)) # the label locations
width = 0.3 # the width of the bars
strLabel1 = 'Neighbor=' + str(round(neighbor, 2))
strLabel2 = 'Neighbor=' + str(round(neighbor+2, 2))
strLabel3 = 'Neighbor=' + str(round(neighbor+3, 2))
axisWidget.axis1.clear()
rects1 = axisWidget.axis1.bar(x - width/2, Neighbor1, width, \
label=strLabel1)
rects2 = axisWidget.axis1.bar(x + width/2, Neighbor2, width, \
label=strLabel2)
rects3 = axisWidget.axis1.bar(x + 3*width/2, Neighbor3, width, \
label=strLabel3)
# Add some text for labels, title and custom x-axis tick labels, etc.
axisWidget.axis1.set_ylabel('Accuracy(%)')
axisWidget.axis1.set_xlabel('Data Ratio (DR)')
axisWidget.axis1.set_title('Accuracy by data ratio (DR) and Number of Neighbors')
axisWidget.axis1.set_xticks(x)
axisWidget.axis1.set_xticklabels(labels)
axisWidget.axis1.legend()
self.autolabel(rects1,axisWidget.axis1)
self.autolabel(rects2,axisWidget.axis1)
self.autolabel(rects3,axisWidget.axis1)
axisWidget.draw()
def graph_DT(self,axisWidget,func):
ratio = self.dsbRatio.value()
depth = self.sbDepth.value()
if (ratio+0.4) < 1 :
rangeDR = [ratio,ratio+0.1,ratio+0.2,ratio+0.3,ratio+0.4]
else :
rangeDR = [ratio-0.4,ratio-0.3,ratio-0.2,ratio-0.1,ratio]
labels = [str(round(rangeDR[0],2)), str(round(rangeDR[1],2)), \
str(round(rangeDR[2],2)), str(round(rangeDR[3],2)), \
str(round(rangeDR[4],2))]
Depth1 = []
for i in rangeDR:
acc = func(i,depth)
Depth1.append(acc)
Depth2 = []
for i in rangeDR:
acc = func(i,depth+4)
Depth2.append(acc)
Depth3 = []
for i in rangeDR:
acc = func(i,depth+4)
Depth3.append(acc)
x = np.arange(len(labels)) # the label locations
width = 0.3 # the width of the bars
strLabel1 = 'Depth=' + str(round(depth, 2))
strLabel2 = 'Depth=' + str(round(depth+2, 2))
strLabel3 = 'Depth=' + str(round(depth+4, 2))
axisWidget.axis1.clear()
rects1 = axisWidget.axis1.bar(x - width/2, Depth1, \
width, label=strLabel1)
rects2 = axisWidget.axis1.bar(x + width/2, Depth2, \
width, label=strLabel2)
rects3 = axisWidget.axis1.bar(x + 3*width/2, Depth3, \
width, label=strLabel3)
# Add some text for labels, title and custom x-axis tick labels, etc.
axisWidget.axis1.set_ylabel('Accuracy(%)')
axisWidget.axis1.set_xlabel('Data Ratio (DR)')
axisWidget.axis1.set_title('Accuracy by data ratio (DR) and Depth')
axisWidget.axis1.set_xticks(x)
axisWidget.axis1.set_xticklabels(labels)
axisWidget.axis1.legend()
axisWidget.axis1.set_facecolor('xkcd:light yellow')
self.autolabel(rects1,axisWidget.axis1)
self.autolabel(rects2,axisWidget.axis1)
self.autolabel(rects3,axisWidget.axis1)
axisWidget.draw()
def graph_LR(self,axisWidget,func):
ratio = self.dsbRatio.value()
learningRate = self.dsbRate.value()
if (ratio+0.4) < 1 :
rangeDR = [ratio,ratio+0.1,ratio+0.2,ratio+0.3,ratio+0.4]
else :
rangeDR = [ratio-0.4,ratio-0.3,ratio-0.2,ratio-0.1,ratio]
labels = [str(round(rangeDR[0],2)), str(round(rangeDR[1],2)), \
str(round(rangeDR[2],2)), str(round(rangeDR[3],2)), \
str(round(rangeDR[4],2))]
LR01 = []
for i in rangeDR:
acc = func(i,learningRate)
LR01.append(acc)
LR001 = []
for i in rangeDR:
acc = func(i,learningRate+0.1)
LR001.append(acc)
LR0001 = []
for i in rangeDR:
acc = func(i,learningRate+0.25)
LR0001.append(acc)
x = np.arange(len(labels)) # the label locations
width = 0.3 # the width of the bars
strLabel1 = 'LR=' + str(round(learningRate, 2))
strLabel2 = 'LR=' + str(round(learningRate+0.1, 2))
strLabel3 = 'LR=' + str(round(learningRate+0.25, 2))
axisWidget.axis1.clear()
rects1 = axisWidget.axis1.bar(x - width/2, LR01, \
width, label=strLabel1)
rects2 = axisWidget.axis1.bar(x + width/2, LR001, \
width, label=strLabel2)
rects3 = axisWidget.axis1.bar(x + 3*width/2, LR0001, \
width, label=strLabel3)
# Add some text for labels, title and custom x-axis tick labels, etc.
axisWidget.axis1.set_ylabel('Accuracy(%)')
axisWidget.axis1.set_xlabel('Data Ratio (DR)')
axisWidget.axis1.set_title('Accuracy by data ratio (DR) and learning rate (LR)')
axisWidget.axis1.set_xticks(x)
axisWidget.axis1.set_xticklabels(labels)
axisWidget.axis1.legend()
axisWidget.axis1.set_facecolor('xkcd:light yellow')
self.autolabel(rects1,axisWidget.axis1)
self.autolabel(rects2,axisWidget.axis1)
self.autolabel(rects3,axisWidget.axis1)
axisWidget.draw()
def autolabel(self,rects,axisWidget):
"""Attach a text label above each bar in *rects*, displaying its height."""
for rect in rects:
height = rect.get_height()
axisWidget.annotate('{}'.format(height),
xy=(rect.get_x() + rect.get_width() / 2, height),
xytext=(0, 3), # 3 points vertical offset
textcoords="offset points",
ha='center', va='bottom')
if __name__ == '__main__':
import sys
app = QApplication(sys.argv)
ex = DemoGUIScikitFeature()
ex.show()
sys.exit(app.exec_())