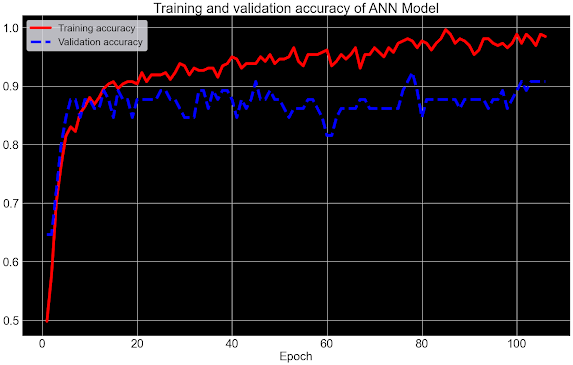
ANN MODEL:
The code implements early stopping, a technique used during model training to prevent overfitting. The EarlyStopping callback monitors the validation loss (val_loss) during training and halts the training process if the loss does not improve over a specified number of epochs (patience). The restore_best_weights parameter ensures that the model's weights are restored to the point where the validation loss was at its minimum. This helps prevent overfitting and improve generalization. Next, TensorFlow is imported, and a sequential model (ann) is initialized using the Sequential class. This model will be used to build the artificial neural network (ANN) architecture.
The architecture of the artificial neural network is constructed step by step using the add() method. The input layer is defined with 1000 units (neurons), corresponding to the number of input features (12 in this case). It uses the ReLU activation function, known for its ability to introduce non-linearity. A dropout layer with a rate of 0.5 is added to mitigate overfitting by randomly deactivating a fraction of neurons during each training batch. Three hidden layers follow: a layer with 500 units, a layer with 250 units, and a layer with 125 units. Each hidden layer employs the ReLU activation function and a dropout layer for regularization. Finally, an output layer with a single neuron and a sigmoid activation function is added to produce binary classification predictions.
After constructing the architecture, the model is compiled using the Adam optimizer and the binary cross-entropy loss function, suitable for binary classification tasks. The model is configured to measure the accuracy metric during training. The EarlyStopping callback created earlier is incorporated into the training process, ensuring that training stops early if the validation loss doesn't improve for a specified number of epochs. The fit method is then used to train the model using the training data (X_train and y_train) with batch size 64. The training data is split into a training set and a validation set (20%) for monitoring the model's performance during training.
Once training is complete, the trained model is saved in the 'heart_model.h5' file using the save method. Additionally, the training history, which contains metrics such as loss and accuracy recorded during each epoch, is saved in the 'heart_history.npy' file using NumPy's np.save function. This history can be later used for visualization and analysis.
In summary, the code demonstrates the construction, compilation, training, and saving of an artificial neural network (ANN) for binary classification using TensorFlow and Keras. The inclusion of early stopping enhances the model's ability to prevent overfitting, while the sequential architecture, dropout layers, and activation functions contribute to building an effective and regularized ANN. The saved model and training history facilitate future analysis, visualization, and potential deployment for making predictions related to heart failure outcomes.
#heart_failure_deep_learning.py import numpy as np # linear algebra import pandas as pd # data processing, CSV file I/O import os import cv2 import pandas as pd import seaborn as sns sns.set_style('darkgrid') from matplotlib import pyplot as plt from sklearn.preprocessing import LabelEncoder from sklearn.model_selection import train_test_split from sklearn.preprocessing import StandardScaler import tensorflow as tf from sklearn.metrics import confusion_matrix, classification_report, accuracy_score from imblearn.over_sampling import SMOTE from tensorflow.keras.callbacks import EarlyStopping def splitt_dataset(): #Reads dataset curr_path = os.getcwd() df = pd.read_csv(curr_path+"/heart_failure_clinical_records_dataset.csv") X = df.drop(['DEATH_EVENT'],axis=1) y = df['DEATH_EVENT'] sm = SMOTE(random_state=42) X,y = sm.fit_resample(X, y.ravel()) #Splits the data into training and testing X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.2, random_state = 2021, stratify=y) #Standar scaler sc = StandardScaler() X_train = sc.fit_transform(X_train) X_test = sc.transform(X_test) return X_train, X_test, y_train, y_test def plot_confusion_matrix(y_test, y_pred, name): #Confusion matrix: conf_mat = confusion_matrix(y_true=y_test, y_pred = y_pred) class_list = ['DECEASED', 'ALIVE'] fig, ax = plt.subplots(figsize=(25, 15)) sns.heatmap(conf_mat, annot=True, ax = ax, cmap='Greens', fmt='g', annot_kws={"size": 25}) ax.set_xlabel('Predicted labels', fontsize=30) ax.set_ylabel('True labels', fontsize=30) plt.xticks(fontsize=20) plt.yticks(fontsize=20) ax.set_title('Confusion Matrix of ' + name, fontsize=30) ax.xaxis.set_ticklabels(class_list), ax.yaxis.set_ticklabels(class_list) def plot_real_pred_val(y_test, ypred, name): plt.figure(figsize=(25,15)) acc=accuracy_score(y_test,ypred) plt.scatter(range(len(ypred)), ypred, color="green",lw=5,label="Predicted", s=200, alpha=0.8) plt.scatter(range(len(y_test)), y_test, color="orange",label="Actual", s=200, alpha=0.8) plt.title("Predicted Values vs True Values of " + name, fontsize=30) plt.xlabel("Accuracy: " + str(round((acc*100),3)) + "%", fontsize=25) plt.legend(fontsize=25) plt.xticks(fontsize=15) plt.yticks(fontsize=15) plt.grid(True, alpha=0.75, lw=1, ls='-.') plt.gca().set_facecolor('black') # Set background color plt.show() def plot_accuracy(history, name): acc = history['accuracy'] val_acc = history['val_accuracy'] epochs = range(1, len(acc) + 1) #accuracy fig, ax = plt.subplots(figsize=(25, 15)) plt.plot(epochs, acc, 'r', label='Training accuracy', lw=7) plt.plot(epochs, val_acc, 'b--', label='Validation accuracy', lw=7) plt.title('Training and validation accuracy of ' + name, fontsize=35) plt.legend(fontsize=25) ax.set_xlabel("Epoch", fontsize=30) ax.tick_params(labelsize=30) plt.gca().set_facecolor('black') # Set background color plt.show() def plot_loss(history, name): loss = history['loss'] val_loss = history['val_loss'] epochs = range(1, len(loss) + 1) #loss fig, ax = plt.subplots(figsize=(25, 15)) plt.plot(epochs, loss, 'r', label='Training loss', lw=7) plt.plot(epochs, val_loss, 'b--', label='Validation loss', lw=7) plt.title('Training and validation loss of ' + name, fontsize=35) plt.legend(fontsize=25) ax.set_xlabel("Epoch", fontsize=30) ax.tick_params(labelsize=30) plt.gca().set_facecolor('black') # Set background color plt.show() def prediction(model, X_test, y_test, name): #Sets the threshold for the predictions. In this case, the threshold is 0.5 (this value can be modified). #prediction on test set y_pred = model.predict(X_test) y_pred = [int(p>=0.5) for p in y_pred] print(y_pred) #Performance Evaluation - Accuracy and Classification Report #Accuracy Score print ('Accuracy Score : ' + name, accuracy_score(y_pred, y_test, normalize=True), '\n') #precision, recall report print ('Classification Report : ' + name + '\n',classification_report(y_pred, y_test)) return y_pred def build_train_ann(X_train, y_train): #Creates early stopping early_stopping = EarlyStopping(monitor='val_loss', patience=10, restore_best_weights=True) #Imports Tensorflow and create a Sequential Model to add layer for the ANN ann = tf.keras.models.Sequential() #Input layer ann.add(tf.keras.layers.Dense(units=1000, input_dim=12, kernel_initializer='uniform', activation='relu')) ann.add(tf.keras.layers.Dropout(0.5)) #Hidden layer 1 ann.add(tf.keras.layers.Dense(units=500, kernel_initializer='uniform', activation='relu')) ann.add(tf.keras.layers.Dropout(0.5)) # Hidden layer 2 ann.add(tf.keras.layers.Dense(units=250, kernel_initializer='uniform', activation='relu')) ann.add(tf.keras.layers.Dropout(0.5)) # Hidden layer 3 ann.add(tf.keras.layers.Dense(units=125, kernel_initializer='uniform', activation='relu')) ann.add(tf.keras.layers.Dropout(0.5)) #Output layer ann.add(tf.keras.layers.Dense(units=1, kernel_initializer='uniform', activation='sigmoid')) print(ann.summary()) #for showing the structure and parameters #Compiles the ANN using ADAM optimizer. ann.compile(optimizer = 'adam', loss = 'binary_crossentropy', metrics = ['accuracy']) # Create EarlyStopping callback early_stopping = EarlyStopping(monitor='val_loss', patience=100, restore_best_weights=True) # Train the ANN with early stopping. history = ann.fit(X_train, y_train, batch_size=64, validation_split=0.20, epochs=1000, shuffle=True, callbacks=[early_stopping]) #Saves model ann.save('heart_model_ann.h5') #Saves history into npy file np.save('heart_history_ann.npy', history.history) print (history.history.keys()) def implement_ann_model(): #Reads dataset X_train, X_test, y_train, y_test = splitt_dataset() #Builds and trains ANN model build_train_ann(X_train, y_train) #Loads ANN model ann_model = tf.keras.models.load_model('heart_model_ann.h5') #Gets predicted values y_pred = prediction(ann_model, X_test, y_test, 'ANN Model') #Plots confusion matrix plot_confusion_matrix(y_test, y_pred, 'ANN Model') #Plots true values versus predicted values diagram plot_real_pred_val(y_test, y_pred, 'ANN Model') # Load the saved training history from 'heart_history.npy' history = np.load('heart_history_ann.npy', allow_pickle=True).item() #Plots accuracy plot_accuracy(history, 'ANN Model') #Plots loss plot_loss(history, 'ANN Model') #Implements ANN Model implement_ann_model()
LSTM Model:
The code outlines the detailed implementation of an advanced Long Short-Term Memory (LSTM) model for predicting occurrences of heart failure. The process involves data preprocessing, model architecture design, training, evaluation, and visualization. Here's a step-by-step breakdown:
Data Transformation and Model Design: Initially, the training data is reshaped to match the LSTM input format, organized as (sample count, time steps, features). The complex LSTM model is constructed using Keras, comprising three LSTM layers, each followed by a dropout layer to mitigate overfitting. These LSTM layers consist of 128, 64, and 32 units respectively, and a final dense layer using a sigmoid activation function for binary classification. The model's architecture is summarized for inspection.
Model Compilation and Training: The LSTM model is compiled with the Adam optimizer and binary cross-entropy loss function. The accuracy metric is specified for evaluation. The model is trained using the training data, divided into training and validation sets using an 80-20 split. During training, the dataset is shuffled before each epoch to enhance generalization. The model undergoes 250 epochs with a batch size of 64 samples per batch.
Model Persistence and Training History: Upon training completion, the trained LSTM model is saved to a file named 'heart_lstm_model.h5'. The history of training, encompassing accuracy and loss metrics for each epoch, is stored in an npy file named 'heart_history_lstm.npy'. This historical data becomes pivotal for subsequent analysis and performance visualization.
Model Implementation and Assessment: Implementation of the LSTM model involves reading the dataset, building and training the LSTM model, and evaluating its predictive capabilities. The splitt_dataset() function is employed to segregate data into training and testing subsets, while build_train_lstm() orchestrates the construction and training of the LSTM model. The saved model is loaded through Keras, and predictions are generated using reshaped test data. The model's performance is appraised through diverse techniques, including generating a confusion matrix, depicting the contrast between actual and predicted values, and graphing accuracy and loss evolution across epochs.
Insights from Visualization and Conclusion: The conclusive phase entails visualizing the model's performance. The confusion matrix affords insights into the model's classification accuracy for heart failure and non-heart failure instances. The visual comparison between predicted and actual values provides a tangible assessment of the model's alignment with real-world outcomes. Furthermore, accuracy and loss plots offer a clear depiction of the model's convergence and learning progress over epochs. Collectively, these visualizations furnish a holistic understanding of the model's behavior and efficacy, furnishing valuable insights for potential refinements and optimizations.
To sum up, the code meticulously executes the entire process of crafting, training, evaluating, and visually interpreting the outcomes of a sophisticated LSTM model designed to predict occurrences of heart failure. This thorough approach facilitates comprehensive analysis and facilitates well-informed decisions for future enhancements or adjustments to enhance the model's predictive capabilities.
def build_train_lstm(X_train, y_train): # Reshape the data for LSTM input X_train = X_train.reshape(X_train.shape[0], 1, X_train.shape[1]) # Create a more complex LSTM model lstm_model = tf.keras.models.Sequential([ tf.keras.layers.LSTM(units=128, return_sequences=True, input_shape=(X_train.shape[1], X_train.shape[2])), tf.keras.layers.Dropout(0.3), tf.keras.layers.LSTM(units=64, return_sequences=True), tf.keras.layers.Dropout(0.3), tf.keras.layers.LSTM(units=32), tf.keras.layers.Dropout(0.3), tf.keras.layers.Dense(units=1, activation='sigmoid') ]) print(lstm_model.summary()) # Display the model summary # Compile the LSTM model lstm_model.compile(optimizer='adam', loss='binary_crossentropy', metrics=['accuracy']) # Train the LSTM model history = lstm_model.fit(X_train, y_train, batch_size=64, validation_split=0.20, epochs=250, shuffle=True) # Save the LSTM model lstm_model.save('heart_lstm_model.h5') # Saves history into npy file np.save('heart_history_lstm.npy', history.history) def implement_lstm_model(): #Reads dataset X_train, X_test, y_train, y_test = splitt_dataset() #Builds and trains ANN model build_train_lstm(X_train, y_train) #Loads ANN model ann_model = tf.keras.models.load_model('heart_lstm_model.h5') #Gets predicted values X_test = X_test.reshape(X_test.shape[0], 1, X_test.shape[1]) y_pred = prediction(ann_model, X_test, y_test, 'LSTM Model') #Plots confusion matrix plot_confusion_matrix(y_test, y_pred, 'LSTM Model') #Plots true values versus predicted values diagram plot_real_pred_val(y_test, y_pred, 'LSTM Model') # Load the saved training history from 'heart_history_lstm.npy' history = np.load('heart_history_lstm.npy', allow_pickle=True).item() #Plots accuracy plot_accuracy(history, 'LSTM Model') #Plots loss plot_loss(history, 'LSTM Model') #Implements LSTM Model implement_lstm_model()
LSTM Output:
Model: "sequential"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
lstm (LSTM) (None, 1, 128) 72192
_________________________________________________________________
dropout (Dropout) (None, 1, 128) 0
_________________________________________________________________
lstm_1 (LSTM) (None, 1, 64) 49408
_________________________________________________________________
dropout_1 (Dropout) (None, 1, 64) 0
_________________________________________________________________
lstm_2 (LSTM) (None, 32) 12416
_________________________________________________________________
dropout_2 (Dropout) (None, 32) 0
_________________________________________________________________
dense (Dense) (None, 1) 33
=================================================================
Total params: 134,049
Trainable params: 134,049
Non-trainable params: 0
_________________________________________________________________
None
[0, 0, 1, 1, 1, 1, 1, 1, 0, 1, 1, 0, 1, 1, 0, 0, 1, 1, 0, 1, 1, 1, 0, 1, 1, 1, 1, 1, 0, 1, 1, 0, 0, 1, 0, 0, 0, 1, 0, 1, 1, 1, 1, 1, 0, 0, 1, 0, 1, 0, 0, 1, 1, 0, 0, 1, 0, 1, 0, 1, 0, 0, 0, 1, 0, 1, 0, 1, 1, 0, 0, 1, 0, 0, 0, 1, 0, 1, 1, 1, 1, 0]
Accuracy Score : LSTM Model 0.8414634146341463
Classification Report : LSTM Model
precision recall f1-score support
0 0.78 0.89 0.83 36
1 0.90 0.80 0.85 46
accuracy 0.84 82
macro avg 0.84 0.85 0.84 82
weighted avg 0.85 0.84 0.84 82
RNN Model
def build_train_rnn(X_train, y_train): # Reshape the data for RNN input X_train = X_train.reshape(X_train.shape[0], X_train.shape[1], 1) complex_rnn_model = tf.keras.models.Sequential([ tf.keras.layers.SimpleRNN(units=128, activation='relu', return_sequences=True, kernel_regularizer=tf.keras.regularizers.l2(0.001), input_shape=(X_train.shape[1], 1)), tf.keras.layers.Dropout(0.3), tf.keras.layers.SimpleRNN(units=64, activation='relu', return_sequences=True, kernel_regularizer=tf.keras.regularizers.l2(0.001)), tf.keras.layers.Dropout(0.3), tf.keras.layers.SimpleRNN(units=32, activation='relu', kernel_regularizer=tf.keras.regularizers.l2(0.001)), tf.keras.layers.Dropout(0.3), tf.keras.layers.Dense(units=1, activation='sigmoid', kernel_regularizer=tf.keras.regularizers.l2(0.001)) ]) print(complex_rnn_model.summary()) # Compile the complex RNN model complex_rnn_model.compile(optimizer='adam', loss='binary_crossentropy', metrics=['accuracy']) # Train the complex RNN model history = complex_rnn_model.fit(X_train, y_train, batch_size=64, validation_split=0.20, epochs=250, shuffle=True) complex_rnn_model.save('heart_rnn_model.h5') np.save('heart_rnn_history.npy', history.history) def implement_rnn_model(): # Reads dataset X_train, X_test, y_train, y_test = splitt_dataset() # Builds and trains complex RNN model build_train_rnn(X_train, y_train) # Loads complex RNN model complex_rnn_model = tf.keras.models.load_model('heart_rnn_model.h5') # Reshape the test data for RNN input X_test = X_test.reshape(X_test.shape[0], X_test.shape[1], 1) # Gets predicted values y_pred = prediction(complex_rnn_model, X_test, y_test, 'RNN Model') # Plots confusion matrix plot_confusion_matrix(y_test, y_pred, 'RNN Model') # Plots true values versus predicted values diagram plot_real_pred_val(y_test, y_pred, 'RNN Model') # Load the saved training history from 'heart_complex_rnn_history.npy' history = np.load('heart_rnn_history.npy', allow_pickle=True).item() # Plots accuracy plot_accuracy(history, 'RNN Model') # Plots loss plot_loss(history, 'RNN Model')












No comments:
Post a Comment